DNA mismatch repair is a mechanism of repairing DNA by the removal of mismatched nucleotides via MMR proteins. The role of mismatch repair proteins in the prokaryotic DNA helped to study the mismatch repair system in the eukaryotic DNA as well. The mismatch repair system is essential, as it maintains the genome’s stability at the time of DNA duplication and recombination.
The MMR system makes the use of MMR or Mismatch repair proteins to repair both the prokaryotic and eukaryotic DNA type. In the absence of MMR system, some defect arises, which are associated with:
- Genome instability
- A predisposition of certain types of cancer
- Abnormalities in meiosis cell division
- Sterility in the mammalian system
To explain the MMR system, E.coli is the best model organism. We will come to know the definition, mechanism, types, components and functional role of the mismatch repair system in this context.
Content: DNA Mismatch Repair
Definition of DNA Mismatch Repair
DNA mismatch repair refers to the rescue system that conserves the DNA sequences by removing the erroneous mismatched, inserted or deleted bases during the DNA duplication and recombination. Mismatch repair system functions to maintain genomic stability and integrity. It reduces the chances of error by 20-400 fold in the E.coli.
DNA Mismatch Repair Mechanism
Both the prokaryotic and eukaryotic mismatch repair system follows the same pathway to recover the mismatched DNA:
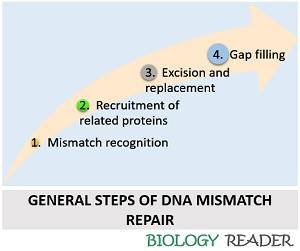
Mismatch Recognition
- In prokaryotic MMR: The MutS protein complex detects the DNA lesion and combines with an ATP molecule, and gets activated.
- In eukaryotic MMR: The MSH protein complex recognizes the non-matched bases in a DNA strand and gets activated by an ATP molecule.
Recruitment of Related Proteins
- In prokaryotic MMR: The activated ATP-MutS complex recruits MutL (acts as a carrier protein). Then, MutS and MutL complex activates the MutH complex. MutS, MutL and MutH form the ternary complex, which loads the DNA helicase-II or UvrD enzyme onto the DNA lesion site.
- In eukaryotic MMR: The activated ATP-MSH complex recruits PCNA and polymerase-δ. Then, ATP dependent conformation change occurs in the mobile clamp. The conformational changes recruit the binding of the MutLa complex. The PCNA (Proliferating cell nuclear antigens) activates the MutLa complex and creates a nick in a daughter strand.
Excision and Replacement
- In prokaryotic MMR: Exonuclease-I is activated by the MutSa protein, which removes the mispaired bases. Then, RDA protein displaces the mismatched base. MutSa and MutLa terminate the exonuclease-I activity.
- In eukaryotic MMR: The MutLa complex promotes the digestion of the mismatched bases by the exonuclease activity-I with the association of RPA (Recognition replication protein A).
Gap Filling
- In prokaryotic MMR: The DNA polymerase III, through its 5’-3’ activity, forms new bases. Single stranded binding proteins (SSB) also bind to the new DNA strand and prevents the new DNA to become double-stranded. Finally, DNA ligase fills the gaps in the DNA strand.
- In eukaryotic MMR: The RFC (Replication factor-C) enzyme plays an important in the synthesis of new DNA bases. Replication factor-C enzyme attaches at the 3’ primer site and activates the DNA polymerase activity to form new bases. Single stranded binding proteins (SSB) prevents the new DNA to become double-stranded. Finally, DNA ligase fills the gap in the DNA strand.
Types of DNA Mismatch Repair
The mismatch repair is generally of two types, namely:
- Prokaryotic MMR
- Eukaryotic MMR
Prokaryotic Mismatch Repair
It consists of four essential components:
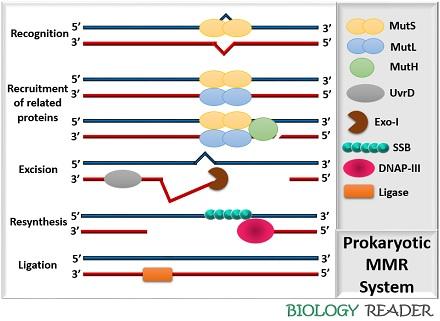
- MutS: It is the most crucial protein complex whose function is to recognize the mismatched bases in the DNA. MutS protein complex is the first enzyme, which initiates the process of MMR by recognizing the non-specific sequences in the DNA. It consists of two specific sites that are sterically distant from each other, as both can affect each other’s function.
- DNA binding site: It helps in associating the MutS protein complex to the DNA lesion site.
- ATPase or Dimerization site: It brings conformational changes in the MutS protein.
- MutL: It acts as an “intermediate protein” complex, which links the MutS protein and endonuclease enzyme. Thus, it is associated with the two activities (recognition and excision of mismatched bases). Firstly, MutL binds with the activated MutS and eventually activates the endonuclease enzyme, i.e. MutH. MutL performs another function by recruiting and loading helicase enzyme (UvrD) at the DNA mismatched site.
- MutH: It belongs to the type-II family of restriction endonucleases. MutH creates a nick in the hemimethylated GATC site. MutS, MutL and an ATP molecule trigger MutH-endonuclease activity. MutH protein also consists of a C-terminal that functions as a molecular attachment site where the other two enzymes (MutS and MutL) communicate and stimulate the MutH activity.
- UvrD: It refers to the “DNA helicase II” enzyme complex, which functions to unwind the DNA lesion site. MutL helps in loading UvrD protein at the mismatched site and stimulates the intrinsic helicase activity of an UvrD enzyme. The SSB proteins and UvrD enters the ss-DNA via nick created by the MutH complex.
- DNA specific exonucleases: Exonuclease enzymes like Exo-I, Exo-X and 5’-3’ exonuclease perform excision of the newly formed DNA between the nick and mismatch.
Eukaryotic Mismatch Repair
It consists of the following components:
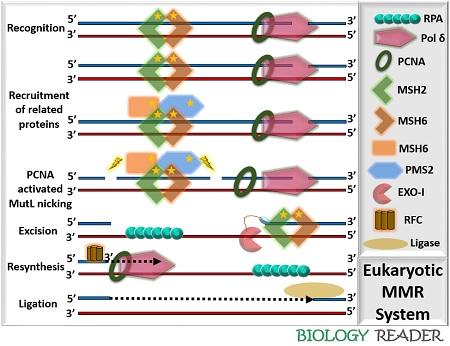
- MSHs protein: It shows homology with the MutS enzyme of prokaryotic MMR. In yeasts and mammals, MSH2 to MSH6 protein complexes are found. MSH enzyme is a heterodimer protein complex, which consists of two domains:
- MutSa: It contains two types of subunits, namely, MSH2 and MSH6. MSH2 contributes 80-90% of the cellular level. MSH6 recognizes the mismatched bases, particularly base-base mismatches and insertion/deletion loops as well.
- MutSb: It contains two types of subunits, namely, MSH2 and MSH3. It mainly repairs the insertion/deletion mispairs.
- MLH protein: It resembles a MutL enzyme of prokaryotic MMR. MLH protein consists of four highly conserved proteins (MLH1, MLH3, PMS1 and PMS2) in yeasts and mammals. It also acts as a heterodimer protein complex and consists of three subunits:
- MutLa: It consists of two subunits MLH1, PMS2. MutLa coordinates with the Mut-S complex to repair the damage.
- MutLb: It consists of two subunits, MLH1, PMS 1. MutLb functions are not known.
- : It consists of two subunits MLH1, MLH3. MutLg repairs an insertion or deletion loop damage.
- Accessory proteins: It includes enzymes like PCNA (Proliferating cell nuclear antigens), Recognition replication protein A (RPA), Replication factor C (RPC), Binding exonuclease-I etc.
Functional Role
DNA mismatch repair involves a simple mechanism of excising mismatched bases along with 3,000 base pairs. Other than the excision of mismatched bases, it also performs some other functions, according to a recent study.
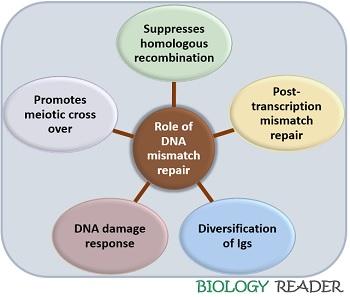
MMR proteins repair the post replication repair of mismatched DNA. Other function like promotion of meiotic cross over, DNA damage response and diversification of immunoglobulins. Suppression of homologous recombination by antirecombinational activity between divergent sequences.