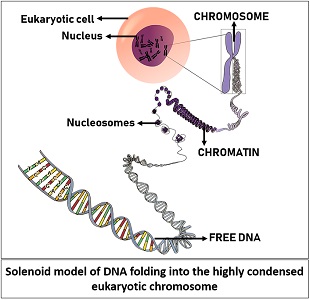
The solenoid model illustrates the packaging or folding of DNA within the eukaryotic cell nucleus. It was first introduced by the scientist’s John Finch and Aaron Klug in the year 1976. In eukaryotes, the DNA is organized compactly due to the association of bead-like histone octamer and linker histones.
Oppositely, the DNA content in prokaryotes is dispersed throughout the cell. The solenoid model is a one-start helix arrangement of nucleosome cores so that each lies parallel to the other along the same helix.
We could easily study the pattern of DNA packaging inside the eukaryotic cell nucleus through electron microscopy and X-ray diffraction. This post describes the definition, important terms, overview and formation of the solenoid model.
Content: Solenoid Model
Definition of Solenoid Model
The solenoid model represents the organization of the nucleosome within the 30 nm wide chromatin fibre. The term solenoid itself defines the winding of the DNA helix. In eukaryotes, the DNA is compressed into chromatin fibres after the association of DNA-linking histone proteins.
The protein complex (histone octamer) attached to the core DNA is termed a nucleosome. Then, the H1 protein seals the wrapped DNA around the core protein. In this way, multiple nucleosomes can polymerize to form oligonucleosome. The nucleosome cores attach through a 35-58 bp long segment of linker DNA, colloquially known as chromatosome.
Important Terms
While studying the solenoid model, we will go through some important terms that we must know.
DNA
It is an acronym for the term deoxyribose sugar. DNA is the hereditary material or fundamental unit of a chromosome. It is composed of nucleosides (a combination of nitrogenous bases and ribose sugar) and a phosphate backbone. The DNA has a complete set of information that plays a humongous role in deciding what a cell should do, how it should look, etc.
Gene
It is the smallest section of DNA within the chromosome. It contains a specific DNA sequence and encodes specific enzymes that regulate genetic attributes, physiology, metabolism, cell replication, etc.
Chromatin
It is the less condensed form of DNA. Chromatin is a macromolecule in which the free eukaryotic DNA twists around the histone octamer that looks like beads on a string. Within the cell nucleus, the DNA condenses into chromatin fibres (30 nm wide) after the DNA packaging around repeating units of nucleosome cores (10 nm wide). Roger Kornberg stated that chromatin fibres appear because of the coiling between the core DNA (200 bp) and repeating units of a core protein complex.
Nucleosome
It is the structural unit of chromatin that merely refers to the repeating globular unit around the core DNA. A nucleosome complex consists of 146 bp long nucleosomal DNA and a core histone complex that possesses eight histone proteins. It appears as a spherical mass that is 11 nm wide with a height of 5.5 nm.
Chromatosome
It refers to the binding of nucleosomes with chromatin fibre. Chromatosome possesses a nucleosome, DNA-linker and H-1 histone proteins. It possesses nine histone proteins, including H1 protein.
Chromosome
It is the highly condensed form of DNA, which becomes visible during the metaphase cell cycle when the cell is ready to divide. It results after the scaffolding of 30 nm chromatin fibres.
Some scaffold attachment regions (A+T rich regions) where the scaffold proteins attach and result in supercoiling of the chromatin fibre with a diameter of 300 nm. Later, the fibres become more condensed into the 700 nm wide chromatids.
The degree of compaction gradually increases with the leading stages of the cell cycle. During metaphase, chromatids become highly condensed to form chromosomes that are having a diameter of 1400 nm.
Overview of Solenoid Model
The solenoid model explains the compaction of the eukaryotic genome within the small spherical nucleus. Firstly, the free DNA condenses into less-compressed chromatin fibres. A chromatin fibre holds the DNA-helix through repeating globular units or histone proteins. Later, the chromatin fibres condense to a higher degree and transform into a highly compressed structure called a chromosome.
When a cell is ready to divide, the chromosome separates the DNA content equally between the two dividing cells. Through the X-ray diffraction method, the DNA organization in the eukaryotic nucleus gives explicit information about the folding of DNA, degree of compactness and the DNA-linking proteins.
Due to the considerable amount of DNA, the DNA must be fixed so that it can reside in the small nucleus. We can look into the consecutive stacking of nucleosomes that form a coiled loop with six nucleosomes per turn through the solenoid model. The term solenoid represents the coiling of ds-DNA in the form of a coiled loop.
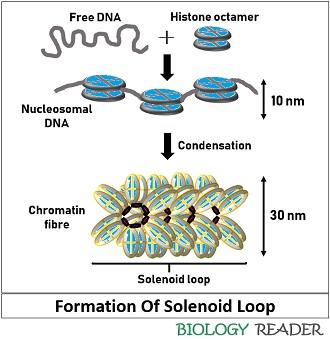
Formation
The solenoid model represents the formation of a helical loop, in which nucleosomes assemble one over the other in a left-handed fashion around the DNA helix. The linker DNA is slightly bent, connecting the nucleosome core with the solenoid or DNA-helix to form a long chromatin fibre.
The association of two subunits, each of H2A, H2B, H3 and H4, form histone octamer. The organization of histone proteins is illustrated in the diagram below.
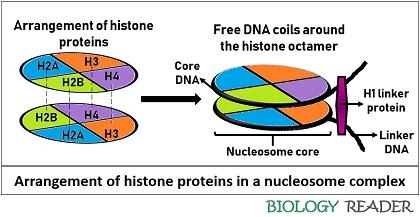
The histone proteins are organized in a way to produce steric hindrance repulsion force. Two factors, namely stereo-specific bond and anionic nucleoplasmin protein, associate histone subunits. Therefore, these two factors stabilize the core histone complex.
Then, the DNA (147 bp) coils around the core protein particle by 1.75 times. In simple words, the DNA takes one full turn and later a 3/4th turn to form a single nucleosome. Finally, H1 linker proteins seal nucleosomal units.
Multiple nucleosomes join linearly, ultimately forming an 11 nm wide nucleosomal fibre that looks like beads on a string. Nucleosomes cores attach through a 20-60 base pairs long segment of linker DNA.
Oligonucleosomes
Multiple nucleosomes combine to form a long nucleosomal fibre of width 10 nm, in which the H1 proteins are located centrally. As a result, H1 proteins interact with each other, which cause H1-H1 proteins repulsion and an electrostatic attraction between the DNA and H1 protein.
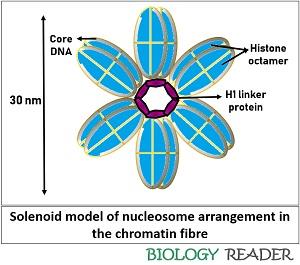
DNA is negatively charged (due to phosphate ions), by which it gets attracted to the positively charged H1 protein (due to the presence of arginine, histidine and lysine residues). There were two models given for the DNA packaging, namely solenoid and zigzag.
In both models, the formation of 30 nm helical fibre from the secondary folding of the 11 nm fibre is common. But, the arrangement of nucleosomes around the helix and the position of linker DNA differs in both models. The solenoid model for DNA packaging was widely accepted by many scientists and is still in use to represent the chromatin organization.
Conclusion
Therefore, a 10 nm wide nucleosomal fibre transforms into a 30 nm wide chromatin in the solenoid model. We can conclude that the nucleosomes are the structural units of chromatin that fix the free DNA into a precise or condensed form within the cell nucleus.
I understood the concept really well.