Tryptophan operon is found within the genome of E.coli, which carries a set of genes constructing an essential amino acid, tryptophan. Sometimes, it is also termed as trp operon. Unlike lactose or lac operon, trp operon is a repressible system discussed in this article.
A scientist named Charles Yanofsky and his co-workers explicitly studied the role of regulatory and structural genes of the trp operon. Trp operon aids biosynthesis of the amino acid tryptophan from a precursor molecule (chorismic acid). Tryptophan functions as an effector molecule that is required for building up the polypeptide chain.
The tryptophan is the end product of the biosynthetic pathway, whose combination or dissociation with the repressor protein can turn on or turn off the trp operon system. This post mainly describes the definition, structure and regulatory system of the tryptophan operon.
Content: Tryptophan Operon
Definition
Trp operon is a repressible system that regulates gene expression for tryptophan biosynthesis according to the binding or uncoupling of a repressor with the operator region. The association or dissociation of the repressor protein strongly depends upon the tryptophan level in the surrounding.
A high level of tryptophan or effector molecule escalates the binding affinity of a repressor protein with the operator sequence, which in turn terminates the gene transcription. In contrast, a low level of tryptophan results in detachment of the repressor from the operator region and allows gene expression.
Structure of Tryptophan Operon
The trp operon consists of:
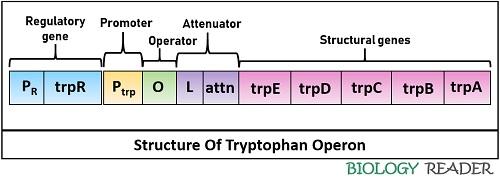
- Promoter (P) region
- Operator (O) region
- Regulatory region
- Attenuator (A) region
- Structural genes (Trp A-E)
Promoter Gene
It refers to the region on a bacterial chromosome that comprises a specific nucleotide sequence, where an RNA polymerase can specifically bind to initiate transcription. The binding of a repressor protein with the operator inhibits the binding of RNA polymerase with the promoter sequence, which in turn terminates the transcription.
Operator Gene
It is the specific nucleotide sequence on chromosomal DNA of E.coli, where a repressor protein can bind by the association of an effector molecule or tryptophan. Here, the tryptophan molecule works as a corepressor that aids activation of aporepressor protein.
Regulatory Gene
Tryptophan operon is a repressor system, in which a regulatory gene of chromosome encodes trp repressor protein that recognizes the operator sequence. The repressor protein switches on the operon system at a low trp level in the surrounding while switches off the system at a high trp concentration in the environment.
Therefore, a repressor protein is associated with the synthesis of five gene products, which depends on the level of tryptophan in the surrounding. A repressor becomes active or inactive in the presence or absence of a corepressor or effector molecule (tryptophan).
Attenuator Region
It is found in the middle of the operator region and structural genes. The attenuator region comprises leader sequences (160 bp in size) that regulate the transcription via attenuation.
Its mechanism is to attenuate transcription efficiency at sufficient tryptophan inside the bacterial cell by forming dimers. Oppositely, the RNAp can traverse through the attenuator region and transcribe genes necessary to build up trp at a low tryptophan level.
Structural Genes
Trp A, B, C, D and E are structural genes of the trp operon. The structural genes encoding for the enzymes and their subunits are necessary for tryptophan biosynthesis from chorismic acid.
- trpE encodes the enzyme Anthranilate synthase I.
- trpD encodes the enzyme Anthranilate synthase II.
- trpC encodes the enzyme N-5’-Phosphoribosyl anthranilate isomerase and Indole-3-glycerolphosphate synthase.
- trpB encodes the enzyme tryptophan synthase-B subunit.
- trpA encodes the enzyme tryptophan synthase-A subunit.
Regulation of Tryptophan Operon
Two mechanisms regulate the trp operon.
- Repressor or derepression mechanism
- Attenuation mechanism
Repression of Tryptophan Operon
It occurs when the trp level is high in the surrounding medium. In this case, the TrpR gene of the tryptophan operon releases apo-repressor (inactive) protein that alone cannot attach to the operator region. In the presence of a corepressor or tryptophan, the apo-repressor protein activates and blocks the RNA polymerase.
Thus, RNAP can not initiate structural genes transcription or inhibits enzymes necessary for the trp construction. Therefore, the respective RNA polymerase can neither bind with the operator gene nor transcribe structural genes at a high tryptophan concentration. Expression of trp operon during availability of tryptophan indicates that the operon system will switch off to terminate the transcription.
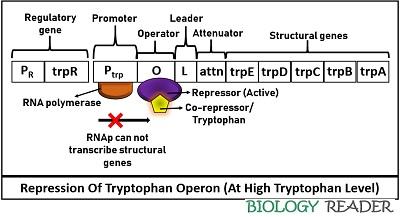
Hence, the repression of trp operon is mediated via complex formed by the association of an allosteric repressor and an effector molecule. Tryptophan functions as a corepressor or an effector molecule that transforms the inactive apo-repressor to an active repressor, which ultimately adheres to the operator sequence to block gene expression.
Derepression of Tryptophan Operon
It occurs in the case of low tryptophan levels in the environment. Due to the lack of sufficient effector molecules, the active repressor protein attached to the operator region will detach. After its dissociation, the repressor remains inactive and functionless. As a result, RNA polymerase becomes free to transcribe structural genes further to produce tryptophan.
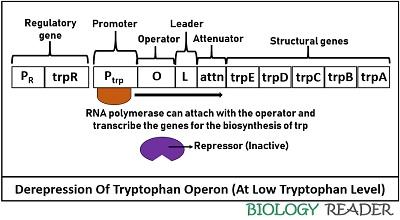
The expression of trp operon during unavailability of tryptophan means that the operon system will switch on to conduct transcription of structural genes by the RNA polymerase. Hence, the derepression is achieved by the dissociation of repressor protein due to lack of trp or effector molecules, which together can form an active complex.
Attenuation
It is the second regulatory region of the trp operon controlled by the trpL gene or attenuator. A leader sequence controls the gene expression via an attenuation mechanism. It comprises a polypeptide sequence plus an attenuator (contains palindromic sequences).
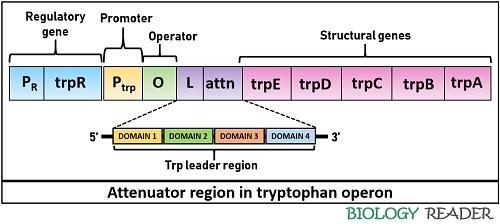
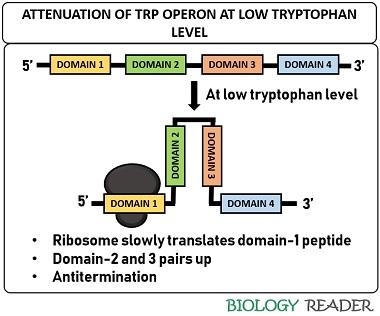
Once the bacterial DNA is transcribed into mRNA, the attenuator sequence can form dimers due to the pairing of palindromic sequences. There are four domains in the leader sequence, in which domain-3 can pair with either domain-2 or domain-4, and domain-1 can pair with domain-2. Besides, it comprises two trp residues.
The pairing of domain-2 and 3 results in antitermination. Conversely, the pairing of domain-3 and 4 causes a termination of trp biosynthesis. The presence of domain-4 (also called attenuator) is important to terminate the transcription because it only can facilitate stem-loop formation. The attenuation mechanism depends upon the pairing of the ribosome and the level of tryptophan inside a bacterial cell.
At low tryptophan level
The ribosome sits at domain-1 of the mRNA transcript. Then, it translates the mRNA very slowly due to the low tryptophan level. As a result, domain-3 interacts with domain-2 due to the halt of the ribosome at domain-1. In such a case, the stem and loop structure will not form. As a result, the transcription may continue to synthesize enzymes necessary for trp production.
At high tryptophan level
The ribosome rapidly translates the domain-1 and sits at the domain-2 when the concentration of tryptophan is high inside the cell. As a result, domain-3 associates with domain-4 and aids in forming a hair-loop structure.
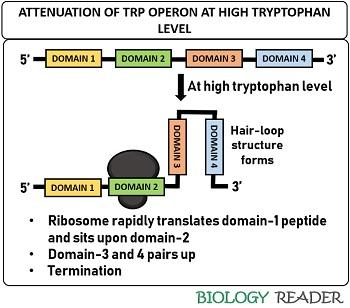
The dimerization of domain-3 and 4 cause the RNAp to fall off and prevent mRNA from transcribing genes encoding enzymes for the trp biosynthesis. Therefore, an attenuator functions as a barrier at high trp concentration due to the pairing of self-complementary sequences.
Conclusion
Therefore, we can conclude that the tryptophan repressor and attenuation system decides when to switch on or switch off the expression of genes synthesizing trp, according to the availability of tryptophan.