DNA polymerase is a complex enzyme. It carries out polymerization of DNA, as it is clear from its name DNA polymerase. Sometimes, it is also called as DNA pol. In prokaryotes, DNA polymerases are typical of three types, namely DNA pol-I, pol-II and pol-III and five types in eukaryotes, namely DNA pol-α, pol-β, pol-Ƴ, Pol- δ and pol-Ɛ.
The first polymerase activity was seen in E.coli, which was observed by Arthur Kornberg in 1958 and he named it as E.coli DNA-pol I. Later, three types of DNA polymerases were introduced and designated as DNA-pol I, DNA-pol II and DNA-pol III. On further discovery, it was found that the DNA polymerase not only have the polymerization activity but also the exonuclease activity.
A DNA polymerase enzyme performs a central role in the DNA duplication process, in which it creates a strand complementary to the template strand in a 5′-3′ direction. Besides, a DNA pol is also involved in the proofreading and editing mechanism of DNA through its exonuclease activity.
In prokaryotes, out of three DNA polymerases, the only one (DNA pol-III) participates in the process of replication and the other two (DNA poly-I and II) perform DNA proofreading and repair. Here, we will discuss all the properties, structure, and activities of all the three polymerases in prokaryotes.
Content: DNA Polymerase
- Definition of DNA Polymerase
- Prokaryotic DNA Polymerases
- Comparison Chart Between the Functions of Prokaryotic DNA Polymerases
- Characteristics
Definition of DNA Polymerase
DNA-polymerase is also called DNA replicate, which forms replicas of DNA relative to the template strand by adding nucleotides to the growing chain. Polymerase enzyme always moves along with the parental strand (template strand) in 3’-5’ direction but forms daughter strand (complementary strand) in 5’-3’ direction. From the name polymerase, we could remember its function that is polymerization or elongation of DNA.
In prokaryotes, there are generally five types of DNA polymerases, out of which three are the most important (DNA pol-I, II and III). DNA pol-I is a key enzyme, which participates in DNA duplication, proofreading, editing, repair and removal of RNA primers. DNA poly-II is solely involved in DNA repair and DNA pol-III is solely involved in the polymerization process. Type IV and V also participates in the DNA repair mechanism but only when the types-I, II and III lose their proofreading efficiency.
Prokaryotic DNA Polymerases
In prokaryotes, DNA pol-I, II and III are the three important types that we will discuss below.
DNA Polymerase I
It is the first polymerase enzyme, which was discovered by Arthur Kornberg in 1958. It consists of a single polypeptide chain. Initially, it was believed that DNA pol-I is a replication enzyme. On further study, it was evidenced that it is more a DNA repair enzyme rather than a replication enzyme. In pol-I, there is one atom of zinc present per chain, due to which it is also known as “Metalloenzymes”.
Activities of DNA pol-I: It shows both polymerases as well as exonuclease activity.
- 5’-3’ polymerase activity involves the addition of nucleotide bases for the synthesis of a new DNA strand.
- 3’-5’ exonuclease activity involves the deletion of mismatched nucleotide bases or it helps in nick translation.
- 5’-3’ exonuclease activity involves the deletion of RNA primers from the 5’end of the complementary DNA strand.
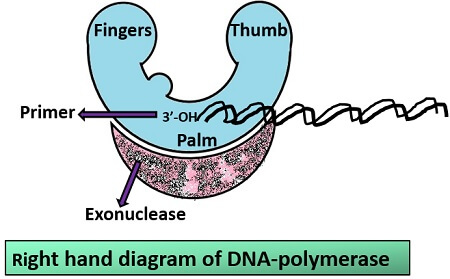
Structure: The structure of Pol-I resembles the right hand of the human. The structure consists of the following three regions:
- Palm region: It is the catalytic active site possessing conserved sequences. It acts as an active site of pol-I. Palm region is made of β- pleated sheet. Its primary function is the processing the addition of deoxyribonucleotide triphosphate.
- Finger region: It is the template site, where once the base pairing is done it encloses the deoxyribonucleotide triphosphate. Its primary function is to catalyze the synthesis of incoming nucleotides by the help of catalyst known as metal ions.
- Thumb region: It is the region which binds the DNA and maintains the correct position of the primer and active site.
DNA Polymerase II
Thomas Kornberg discovered it in 1970. Its efficiency of polymerization is slower than the pol-I. It also consists of a single polypeptide chain. Pol-II acts as backup enzyme or alternative to the process of replication, as in the absence of pol-I it can elongate the Okazaki fragments.
Activities of DNA pol-II:
- 5’-3’ polymerase activity involves the addition of nucleotide bases for the synthesis of a new DNA strand.
- 3’-5’ exonuclease activity involves the deletion of mismatched nucleotide bases or helps in nick translation.
Structure: The structure of pol-II is quite unknown.
DNA Polymerase III
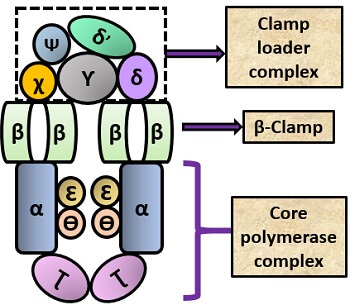
It is the primary holoenzyme that mainly participates in the process of replication. It consists of two polypeptide chains. Pol-III contains subunit of many enzymes that perform different functions, and also called as “heteromultimeric enzyme”. Pol-III contains 10 subunits in its structure that make pol-III a complete enzyme, i.e. “holoenzyme”.
Activities found in DNA pol-III:
- 5’-3’ polymerase activity involves the addition of nucleotide bases for the synthesis of a new DNA strand.
- 3’-5’ exonuclease activity involves the deletion of mismatched nucleotide bases or helps in nick translation.
Structure: The structure of pol-III consists of 10 subunits, such as:
- α: It encodes DNA E gene and helps in DNA synthesis.
- Ɛ: Codes for DNA Q gene and helps in 3’-5’ proofreading activity.
- Ɵ: Encodes hol E gene, acts as an accessory protein and also participates in the proofreading mechanism.
- Ʈ: It codes for DNA X gene and promotes dimerization of the core protein complex.
- Ƴ: It codes for DNA Y gene.
- δ: Encodes hol A gene.
- δ’: Codes for hol B gene.
- χ: encodes hol C gene.
- Ψ: codes for hol D gene.
- β: encodes DNA N gene. It acts as a clamp protein that holds the DNA molecule and responsible for the processivity factor. Ƴ, δ, δ’, χ and Ψ all acts as a clamp loader complex and helps the β- clamp loader protein to bind with the DNA.
Comparison Chart Between the Functions of Prokaryotic DNA Polymerases
| Properties | DNA pol-I | DNA pol- II | DNA pol-III |
|---|---|---|---|
| 5’-3’ polymerase activity | Present | Present | Present |
| 3’-5’ exonuclease activity | Present | Present | Present |
| 5’-3’ exonuclease activity | Present | Absent | Absent |
| RNA dependent polymerization | It can carry out RNA dependent polymerization | It can not | It can not |
| DNA repairing characteristics | Present | Present | Absent |
| Ligation property | Present | Absent | Absent |
| Involvement in replication | Participates | Does not participate | Plays a major role |
Characteristics
DNA- polymerase has some specific characteristics like:
- Polymerization activity
- Fidelity, i.e. proofreading ability
- Processivity, i.e. processing of DNA and its bases
- Thermostability, i.e. stability at high temperature
Example: Taq- DNA polymerase of Thermus aquaticus bacterium.
These all properties make it useful in molecular technologies like PCR, DNA sequencing etc.