Photoreactivation repair is the DNA repair system, which corrects the damage (caused by UV-C and UV-B radiations) with the aid of photolyase enzyme under the exposure of visible light.
The first evidence for the photoreactivation was given by the two scientists, namely Hausser and Von Oehmcke, in 1933. Hausser and Von Oehmcke reversed the darkened banana skin (caused by UV-exposure) by using mercury arc lines at 366, 405 and 435 nm.
Thus, photoreactivation repair is a unique method that repairs the DNA damage-induced due to the light by using light itself. In simple terms, it is a light-dependent process and also called “Light repair”. This lesson will introduce you to the definition, mechanism, role and process of photoreactivation repair.
Content: Photoreactivation Repair
Definition of Photoreactivation Repair
Photoreactivation repair refers to the DNA repair system where the re-exposure repairs the UV-damaged DNA under the visible light. It is also called the direct reversal DNA repair mechanism, in which the photolyase enzyme under the action of visible light reverses the effect of UV-radiation on the cells.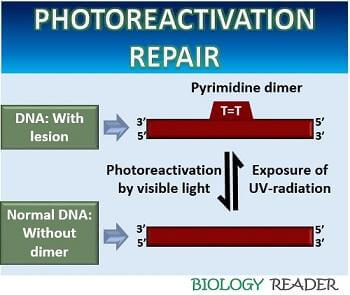
It recovers the DNA from the UV-C (180-290nm) and UV-B (290-320nm) damage by treating it with the light of a longer wavelength (320-500nm). The process of photoreactivation can recover the damage of both prokaryotic and eukaryotic DNA.
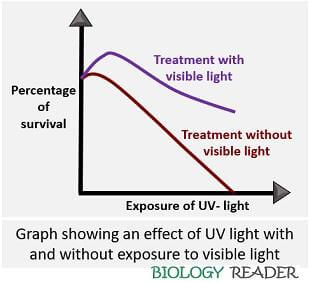
In 1949, Kelner demonstrated photoreactivation by plotting a standard graph between a percentage of survived cells and UV dose. Exponential death with a broad curve was seen in the E.coli cells that were exposed to UV-light. When the E.coli cells were treated with visible light, the size of the slope reduced, as you could see in the graph below:
Mechanism of Photoreactivation
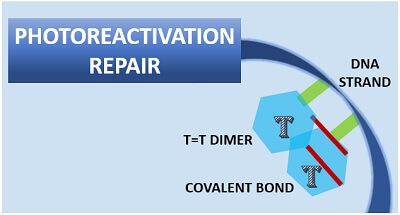
The photoreactivation process repairs the UV-damaged DNA. A DNA helix becomes distorted and forms a pyrimidine dimer (usually thymine=thymine dimer) under UV-light exposure.
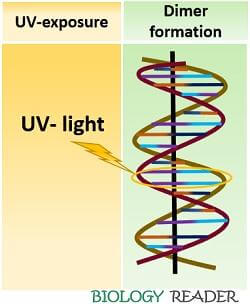
A dimer forms when the adjacent thymine bases attach covalently to their respective C-5 and C-6 atoms. A formation of T=T dimer causes helix distortion, which eventually causes replication errors in the DNA and finally leads to mutation.
The covalent bond formed between the two adjacent thymine bases must be broken to recover the helix distortion. An enzyme photolyase breaks off the covalent bond between the dimer.
Role of Photolyase
The enzyme photolyase consists of FAD (flavin adenine dinucleotide) and MTHF (methylene tetra hydro folate) cofactors.
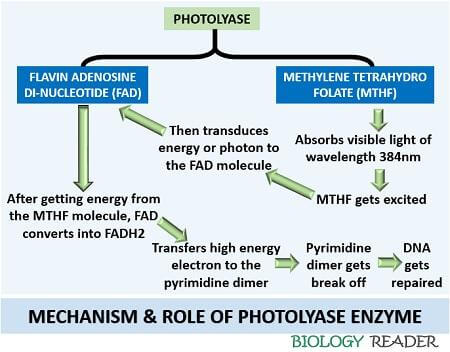
Firstly, the MTHF absorbs visible light of a higher wavelength (Mostly 384nm) to repair the DNA damage. After absorbing the energy coming from the visible light, MTHF gets excited. The excitement of MTHF molecules transduces photons or electrons to the FAD molecule.
Then, the FAD molecule accepts the electron and convert it into FADH2. FADH2 (A high energy molecule) will bind with the pyrimidine dimer. After binding, FADH2 breaks off the double covalent bond between the cyclobutane thymine bases. Thus, a photolyase reverses the cyclobutane pyrimidine bases to the usual bases of the DNA via photochemical reactions.
Process
The process of photoreaction is a light-dependent repair. It uses a photolyase enzyme that activates by getting energy from the longer wavelength’s visible light (320-500nm). Photoreactivation is a direct reversal repair mechanism.
The exposure of visible light to the cyclobutane pyrimidine dimers reverse the effect of UV-exposure. It mainly reverses the dimeric product of UV-light into the usual monomeric form. The process of photoreactivation includes the following steps:
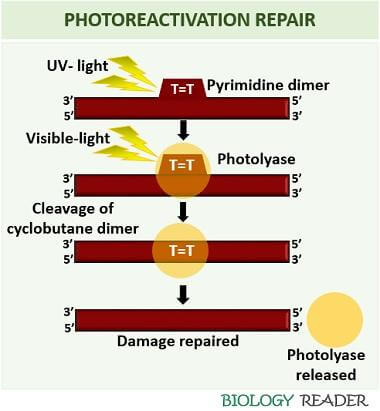
Formation of Pyrimidine-dimer
The UV light induces a covalent bond between the pyrimidine bases (either thymine or cytosine). The two adjacent pyrimidine bases bind through a carbon-carbon double bond. A carbon-carbon double bond formation between the pyrimidine bases forms a dimeric structure commonly referred to as ‘cyclobutane pyrimidine dimers’.
Cyclobutane pyrimidine dimers (CBPBs) is a common product forming under the UV-exposure. In addition to this, 6-4 photoproducts also produce as a result of UV-damage, where the two bases attach with a single carbon bond to the C-6 of one ring and C-4 of another ring. Thymine=Thymine dimer usually forms after the UV-exposure, and it appears like a bubble in the DNA strand.
Treatment with Photolyase under visible light
Under the visible light of a longer wavelength, a photolyase enzyme becomes activated. A photolyase enzyme scans the DNA strand and recognizes the pyrimidine dimers. After recognition of dimeric forms in the DNA, it directly binds to that segment.
Breaking of T=T dimer
When a photolyase enzyme binds to the T=T dimer, it starts splitting the carbon-carbon double covalent bond between the cyclobutane rings. Thus, a photolyase enzyme converts the dimeric form of the pyrimidines bases into monomeric forms.
Release of Photolyase
After breaking the double covalent bond between the consecutive thymine bases, a photolyase enzyme leaves the DNA strand, and the DNA fixes.