Transfer RNA is an RNA type that acts as the intermediary element, and plays a significant role in the loading and transferring amino acids to the site of protein synthesis, i.e. Ribosome. The t-RNA first decodes the information or the nucleotide sequences carried by the m-RNA. Therefore, t-RNA also plays a major role in translating mRNA into proteins.
Let us suppose, the m-RNA carries a code or nucleotide sequences of the DNA and a t-RNA is the key to that code carrying anticodons that can decode the genetic codons of m-RNA. A complementary pairing occurs between the triplet codons of m-RNA with the anticodons of t-RNA.
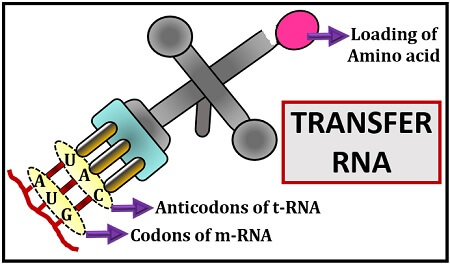
The codons of m-RNA carry genetic information, which decides the type of amino acid that will bind to the amino acid acceptor site of t-RNA. The transfer RNA is named for its key role in transferring the amino acid to the eukaryotic or prokaryotic ribosome after reading the information given by the m-RNA. We will discuss the definition, characteristics, structure (primary, secondary and tertiary) and function of transfer RNA in this context.
Content: Transfer RNA
Definition of Transfer RNA
Transfer RNA is one type of RNA that acts as a carrier molecule that can load the amino acids by decoding the piece of genetic information carried by the m-RNA codons and converting it into the physical proteins. The t-RNA is single-stranded having 5’-3’ end. A letter ‘t’ stands for “Transfer” because tRNA transfers the activated amino acids to the ribosomal site or the site of protein synthesis.
Characteristics of Transfer RNA
- Transfer RNA is also called “Soluble RNA” because it is soluble in the solution of a 1M concentration of NaCl.
- The t-RNA sometimes interchangeable with the term “Adaptor RNA” because it carries the amino acids to the ribosomes.
- Transfer RNA contributes about 10-15% of the cellular RNA.
- The t-RNA is a highly folded structure due to which the nucleotides come closer to each other and appears to be like double-stranded.
- The sedimentation constant of t-RNA is 3.8 S and molecular weight of 25,000–30,000 D.
- The t-RNA is the smallest form of RNA composed of 75-95 nucleotide bases.
- There are twenty types of t-RNA, as there are 20 types of amino acids.
- Transfer RNA is different from the other RNA molecules as it consists of abnormal and modified bases like pseudouracil, inosine, dihydrouridine etc.
- Formation of the stem-loop structure occurs in the t-RNA, due to abnormal base pairs and no hydrogen bonding. In case of regular base pairs and the hydrogen bond between them do not result in such loop formation.
- The DHU arm of t-RNA consists of di-hydroxy uridine as an abnormal base.
- Anticodon arm of t-RNA comprises inosine as an abnormal base.
- The TΨC arm consists of pseudouracil as an unusual nucleotide.
Structure of Transfer RNA
There are three structural configurations of transfer RNA, namely primary, secondary and tertiary structure.
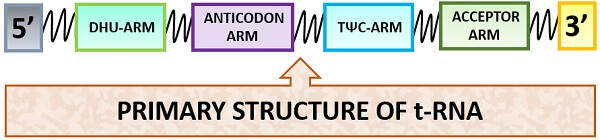
Primary Structure
In the primary structure, the t-RNA possesses a linear structure and consists of 60-90 nucleotides. The primary structure forms “Post-transcriptionally”. On its 3’-end, CCA sequence is present. The charged tRNA assembles the amino acids in correct order where the COOH-group attaches above the 3’-OH end. It is having a large number of modified bases. There are 15 invariant and 8 semi-invariant residues present in the primary structure of t-RNA, which leads to the formation of the secondary and tertiary structure of tRNA.
Secondary Structure
The “Clover-Leaf model” is the most popular model to explain the secondary structure of transfer RNA. Robert Holley gave it in 1968. According to the clover-leaf model, four arms exist in the structure of the t-RNA, and sometimes one additional arm can also be present. Acceptor, DHU, Anticodon and TΨC are the four common arms found in the tRNA structure. In addition to this, one variable arm may also present.
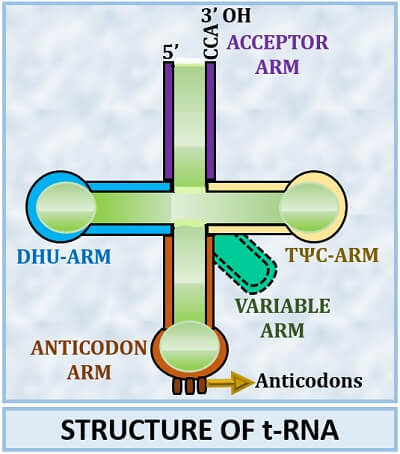
Acceptor’s arm
The “Amino acid attachment site” consists of 7 paired nucleotides and 4 unpaired bases. The acceptor’s arm is a double-helical stem having both 3’ end and 5’ end. The 3’-end consists of a CCA sequence or “Terminal ACC-sequence”. The carboxyl group of amino acid attaches with the 3’–OH end. At the 5’-end, guanine is always present. Therefore, it purely refers to the attachment site for the activated amino acids.
DHU arm
Here, DHU stands for “Di-hydroxy uridine”. The DHU sometimes refers to as “D-arm” that consists of 15-18 total nucleotide bases, from which 7-12 are the modified bases and 4 with usual bases. D-arm comprises of a 4bp long stem and a loop with 7 unusual pyrimidine bases including dihydroxy uridine. D-arm is also known as “Aminoacyl synthetase binding site” as it activates the amino acid synthesis by synthesizing an enzyme (aminoacyl tRNA synthetase).
The enzyme promotes the specific and robust binding of amino acid to the tRNA with the help of an ATP molecule. DHU arm plays a very crucial role in the stabilization of the tRNA tertiary structure. There are two variable regions present on both the sides of guanine residues (α and β variable areas).
Anticodon arm
It is the “Codon recognition site”. Anticodon arm also consists of a stem, which is 5bp long and a loop consist of 7 unpaired bases. In between the seven unpaired bases of anticodon loops, there are three anticodons. The anticodons recognize the codons of m-RNA, and complementarily binds to it.
The anticodons in the loop decide the type of amino acid, which will attach to the 3’-end of the t-RNA. Therefore, the anticodon arm is an essential part of the transfer RNA that recognizes and reads the information carried by the m-RNA.
TΨC arm
Here, TΨC stands for “Thymine pseudouracil cytosine”. TΨC arm also consists of a stem (5bp long) and a loop with 7 unpaired bases. Due to the presence of TΨC sequence in 5’-3’ direction, it is named as TΨC arm. It also possesses a “Ribosomal recognition site” that plays a vital role in binding tRNA with the ribosome.
Variable arm
It is the additional arm, which is present between the anticodon and TΨC arms. The variable arm is the shortest arm whose presence differs among species to species. The length of the variable arm recognizes the enzyme translated for the t-RNA. The variable arm helps in the stabilization of the t-RNA. Based on the presence of a variable arm, t-RNA are of two types:
Tertiary Structure
The tertiary structure of tRNA has a three-dimensional structure that is highly folded and has an L-shaped structure.
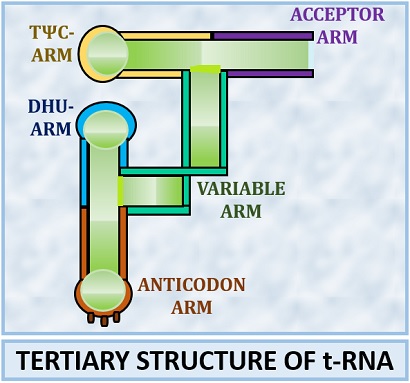
In the tertiary structure of t-RNA, acceptor and TΨC arm form the extended helix downwards. The anticodon and DHU arm form the extended helix upwards. Due to the change in structural configuration, the DHU and TΨC join where both the extended helices align at the angle of 90 degrees. The stabilization of the tertiary structure is through the base pairing and base stacking.
Role of Transfer RNA
The t-RNA first recognizes the codons of mRNA with the help of the anticodon arm. The anticodon arm of t-RNA consists of three anticodons, which then complementarily binds with the codons of the m-RNA. This complementary binding activates the synthesis of an enzyme known as “Aminoacyl tRNA synthetase”. The synthetase enzyme will help in the binding of specific amino acid according to the mRNA specific codons that will code the particular amino acid.
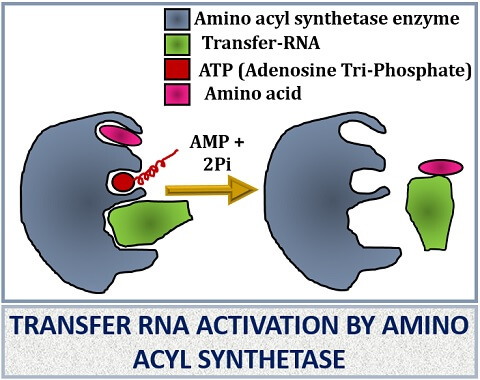
The aminoacyl tRNA synthetase enzyme is different for each amino acids. When both amino acid and the t-RNA molecule attaches with the synthetase enzyme, it will promote both t-RNA and amino acid binding by utilizing an ATP molecule. Once the synthetase enzyme charges the t-RNA, it will load the amino acid and transfer it to the ribosome where the amino acid combines to build up of protein.
Concise and well structured