A restriction enzyme is produced within the bacterial cell due to which it is also called “Restriction endonuclease”. In 1978, Werner Arber, Daniel Nathans, Hamilton O Smith won the Nobel Prize for the characterization and discovery of restriction enzyme during the study of Entero-bacteriophage.
According to their study, the bacteria possess some restriction activity that can inhibit the replication of bacteriophage DNA. Because of the cutting action of the restriction enzymes, they are also called molecular scissors in the recombinant DNA technology or molecular biology. The enzyme catalyses the different reactions and also restricts the function of cleavage.
In this context, we will study the definition, mechanism, nomenclature, types and working of the restriction enzymes. In addition, you will get to know the modified restriction enzyme and the gene involved in the restriction-modification and the modification system only.
Content: Restriction Enzyme
- Definition of Restriction Enzyme
- Mechanism
- Modified Restriction Enzyme
- Nomenclature
- Genes for the Restriction-Modification System
- Genes for the Modification System only
- Types
- Working
Definition of Restriction Enzyme
A restriction enzyme is a type of endonuclease enzyme, which functions to cleave the nucleotide sequences in between the DNA strand. But the site of cleavage is specific for the restriction endonuclease. In the DNA, there are some specific sequences termed as “Recognition or Restriction sequences”. The restriction endonuclease will recognize the restriction sequences, bind to the site and eventually cleave the DNA strand (particularly on restriction sequences) into two or more fragments.
A restriction endonuclease can perform three functions like recognition of restriction site, cleavage in the restriction site and modification of DNA. The restriction endonuclease can be isolated from the bacteria and can be used in genetic engineering and cloning methods etc.
Mechanism of Restriction Enzyme
Let us suppose a bacterial cell infected by phage particle. Then, we could see in the diagram that the phage genome will enter into the bacterial genome. After that, a war begins between a genome of both bacteria and the phage. Both will produce a restriction endonuclease as a weapon to degrade each other. The bacterial genome will produce a restriction enzyme to protect its cell machinery being hijacked by the bacteriophages and also for the degeneration of the phage DNA.
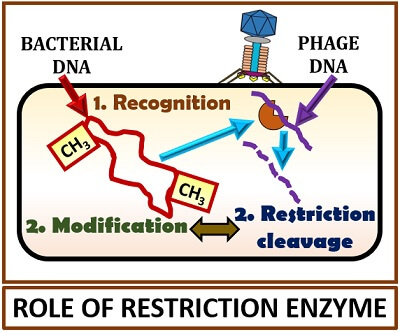
In counter to bacterial’s restriction endonuclease, the phage DNA will produce a restriction endonuclease to degrade the bacterial chromosome by integrating its viral genome. Thus, the recognition sites or sequences present on the bacterial DNA will produce restriction endonucleases, which in turn cleave the foreign DNA at the specific site.
Modified Restriction Enzyme
It is also called “Methylation enzyme”. Modified restriction endonuclease plays a crucial role in the recognition or differentiation of the bacterial vs foreign DNA. In the modified restriction endonuclease, the recognition sequences of the DNA are modified by the methylation. Let us suppose, there is a recognition sequence of “GATC”, then the CH3 or methyl group will attach to the adenine and cytosine thereby modifying the bases.
The modification of the bases guides the restriction endonuclease not to cleave the DNA of its own strand. Modified restriction endonuclease is absent in the viral genomic DNA. This enzyme can either work independently or coordinate with the restriction endonuclease. Therefore, the bacteria will produce modifying restriction endonuclease in order to not cleave its own DNA.
Nomenclature of Restriction Enzyme
First, the restriction enzyme was isolated from the bacteria and the nomenclature system depends upon the type of bacteria from which the enzyme has been isolated. Let us take some examples to understand the nomenclature system.
-
- Example 1 (Eco R-I): The first letter is always written in the capital letter where ‘E’ represents the “Genus” of the bacteria from which the restriction endonuclease has been isolated, i.e. Escherichia. The second two letters represent the species of the bacteria, which is coli and written as ‘co’. The third letter ‘R’ represents the strain of the bacteria which varies with different species. The last is the number, i.e. ‘I’ which signifies the order of identification of the enzyme. The recognition sequence of Eco R-I is 5’-GAATTC-3’.
- Example 2 (Bam H-I): Similarly, the letter ‘B’ represents the genus “Bacillus”. The letters ‘am’ represent the species that is amyloliquefaciens. And, the last two represents the strain number and the order of identification of the restriction endonuclease. The recognition sequence of Bam H-I is 5’-GGATCC-3’.
Genes for the Restriction-Modification System
The genes for the restriction-modification system was first discovered in the E.coli of strain K-12. After the discovery, the genes which are involved in the modification system of restriction endonuclease named as “hsd”. The hsd stands for “Host-specific defence or Host specificity determinant”. The hsd genes in the E.coli of strain K-12, perform three functions, according to which they are classified into three types:
- hsd-S: It includes the specificity factors, i.e. restriction site or recognition sequences, which help the restriction endonuclease to recognize and bind with the specific site of the DNA sequence.
- hsd-M: It includes the modification factors, which modify the nucleotide bases of the DNA by adding CH3 group that prevent the chopping up of its own DNA by the restriction endonuclease.
- hsd-R: It codes for the particular site or sequences in the DNA, where the restriction endonuclease binds and cleaves the strand into two or more fragments.
Genes for the Modification System only
The modified restriction enzyme can also work independently. The genes which control the modified restriction enzyme are of three types:
- dam gene: Here, the letters dam stands for “DNA Adenine Methyltransferase”. The dam gene recognizes particularly 5’-GATC-3’ sequence and the methyl group attaches to the adenine base.
- dcm gene: Here, the letters dcm stands for “DNA Cytosine Methyltransferase”. The dcm gene recognizes particularly 5’-CCATGG-3’ sequence and the methyl group attaches to the cytosine base.
- mcr gene: Here, letters mcr stands for “Modified Cytosine Restriction”. The mcr genes are of two types, namely mcr B and mcr C. It recognizes the modified sequences and cut the DNA from the internal cytosine residue.
Types of Restriction Endonuclease
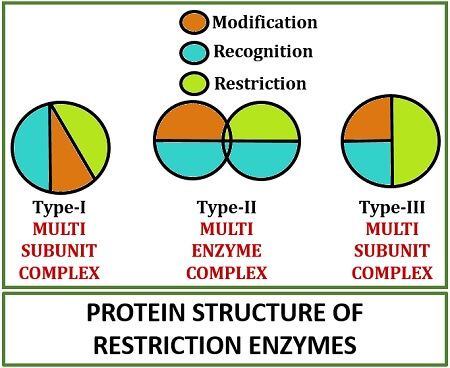
The restriction endonuclease is classified into three types namely type-I, II and III, based on the following factors like enzyme system complexity, cofactor requirements etc. There is a comparison table given below that differentiates all the three types of restriction enzymes.
| Properties | Type-I | Type-II | Type-III |
|---|---|---|---|
| Protein structure | Multi-subunit complex | Multi-enzyme complex | Multi-subunit complex |
| Cleavage site | 1000bp downstream | At the recognition site | 24-26bp downstream |
| Requirements of DNA cleavage | Two recognition sequences, in any orientation | One recognition sequence | Two recognition sequences, in and out direction |
| Cofactors requirement | ATP, Mg2+ and sodium adenosyl methionine | Mg2+ | ATP, Mg2+ and sodium adenosyl methionine |
| Site of methylation | At the recognition site | At the recognition site | At the recognition site |
| Importance in cloning | Not preferred for the cloning | Preferred enzyme for the cloning and genetic engineering | Not preferred for the cloning |
Type-I Restriction endonuclease: The type-I restriction endonuclease is a single enzyme, but acts as a “multi-subunit complex”. This enzyme creates a cleavage at 1000bp downstream to the DNA strand. Type-I restriction endonuclease requires two recognition sequences in any orientation. For the cleavage, it requires ATP, Mg2+ and sodium adenosylmethionine. In the process of cloning, it is not preferred because of the non-specificity of the cleavage site.
Type-II Restriction endonuclease: The type-II restriction endonuclease is a set of an enzyme that acts as a “multienzyme complex”. This enzyme creates a cleavage specifically on the recognition site of the DNA strand. Type-II restriction endonuclease requires one recognition sequence. For the cleavage, it requires Mg2+. In the process of cloning, it is the most preferred enzyme because of its specificity in the cleavage site.
Type-III Restriction endonuclease: The type-III restriction endonuclease is a single enzyme with a “multi-subunit complex”. This enzyme creates a cleavage at 24-26bp downstream to the DNA strand. Type-I restriction endonuclease requires two recognition sequences to read the DNA strand in the bi-directional movement facing towards each other.
For the cleavage, it requires ATP, Mg2+ and sodium adenosylmethionine. In the process of cloning, it is not preferred because of the non-specificity of the cleavage site where one has to count the 26bp downstream to cut the DNA strand. The site of methylation occurs at the recognition site.
Working of Restriction Endonuclease
As the restriction endonuclease can perform either of the two functions like cleavage or modification. Both the processes cannot occur simultaneously so a restriction enzyme has to decide when it has to modify or to cleave the DNA strand.
Case-1: In bacterial DNA
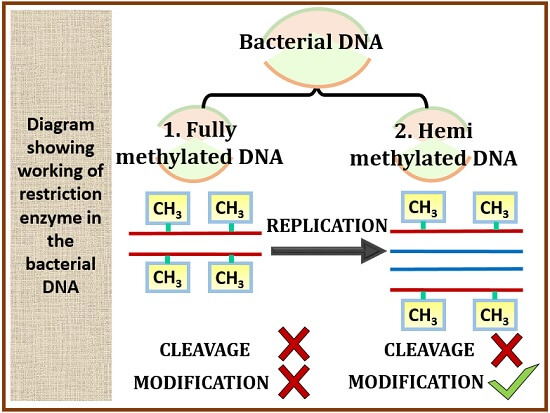
When the bacterial DNA is having fully methylated DNA strand, the restriction endonuclease and the modified restriction enzyme neither participate in cleavage nor modification. Cleavage will not occur in this case, because both the strands of DNA are methylated so it will not allow the restriction endonuclease to cut its own strand. Likewise, the modified restriction enzyme cannot do any modifications in case of fully methylated DNA.
But in hemimethylated DNA, only one strand of the DNA is methylated. Fully methylated DNA when replicate, it will produce hemimethylated DNA. Therefore, in hemimethylated DNA, new strands of non-methylated DNA will produce. The modified restriction endonuclease can also modify the new strands of the hemimethylated DNA and the restriction endonuclease will not work in case of methylated DNA.
Case-2: In phage DNA
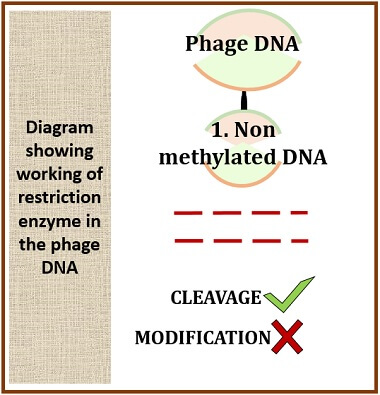
In a foreign cell, the DNA is non-methylated. In this case, the modified restriction enzyme will not work because the modification system is not found in the foreign cell. As there is no modification or methylation, the restriction endonuclease will cut the DNA strand into some fragments.