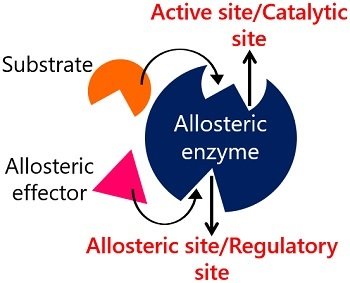
The allosteric site of an enzyme is regulatory in function. Enzymes with allosteric or regulatory sites behave as the “Allosteric enzymes”. Such enzymes possess multiple binding sites for both substrates and effectors. Allosteric enzymes are regulated by the non-covalent or reversible binding of effectors or non-substrate molecules to the allosteric site.
Thus, the allosteric site allows the effector molecules to ‘switch on’ or ‘switch off’ of the enzyme activity. An allosteric or regulatory site is a place where the effectors bind and they may function as the allosteric activators or inhibitors. We can call effectors the modulators because they alter the enzyme’s configuration to promote or obstruct the substrate binding to the enzyme’s active site.
This post mainly describes the definition of allosteric enzymes and allosteric sites along with some key features. In addition, you will get to know the allosteric enzymes examples, types and models of allosteric regulation as well as the kinetics of allosteric enzymes.
Content: Allosteric Site
- Definition of Allosteric Enzyme
- Definition of Allosteric Site
- Features
- Allosteric Enzymes Examples
- Regulation
- Models
- Sigmoidal Curve
- Conclusion
Definition of Allosteric Enzyme
It is the oligomeric enzyme that possesses multiple protein subunits. In simple words, enzymes with an extra or allosteric site other than the active site are called allosteric enzymes. The activity of allosteric enzymes depends upon the type of effector or modulator that binds to the allosteric site. Allosteric enzymes appear larger and structurally more complex than simple or non-allosteric enzymes.
Definition of Allosteric Site
It refers to the regulatory site of an enzyme, which provides a binding site for the effector or non-substrate molecules that either activate or inhibit the enzyme’s catalytic efficiency. The interaction between effectors and the enzyme’s allosteric or regulatory site changes the overall shape of the enzyme, either enabling or preventing substrate binding to the enzyme’s catalytic site.
Features
- Allosteric enzymes have multiple protein subunits or possess many polypeptide chains, due to which they exist in a quaternary shape.
- They share a few common characteristics similar to simple enzymes. However, they follow a sigmoidal curve for a reaction rate vs substrate concentration instead of a hyperbolic curve.
- The allosteric site is physically disparated from the active site of an enzyme.
- Allosteric enzymes exist in the active form (R-state) or inactive form (T-state).
- They often have multiple binding sites for substrates and effectors.
- Allosteric enzymes carry a catalytic and regulatory site. A catalytic site serves as an active site, and a regulatory site serves as an allosteric site.
Allosteric Enzymes Examples
Let us discuss some common examples of regulatory enzymes.
- Phosphofructokinase is a regulatory enzyme of the glycolytic pathway, having two active sites. One is for substrate binding, while the other functions as a regulatory site promoting ATP or AMP binding. The binding of AMP enhances the catalytic efficiency of phosphofructokinase or stimulates glycolysis. Oppositely, the binding of ATP allosterically inhibits the activity of PFK or ceases glycolysis.
- Aspartate transcarbamoylase or ATCase is also a regulatory enzyme of the pyrimidine biosynthetic pathway, possessing a large catalytic subunit and a smaller regulatory subunit. The interaction of a regulatory subunit with the final product (cytidine triphosphate) inhibit ATCase activity.
Allosteric Enzyme Regulation
Allosteric modulators or effectors regulate the activity of allosteric enzymes. An allosteric effector serves as a metabolite that binds to the allosteric site, causing conformational or electrostatic changes resulting in stimulation or inhibition in the enzyme’s activity.
- Oxygen is a homotropic effector of haemoglobin.
- Hydrogen, carbon dioxide and 2,3-bisphosphoglycerate are the heterotropic effectors of haemoglobin.
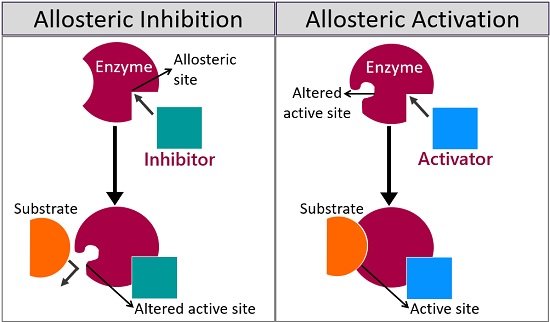
Allosteric activators enhance the enzyme’s activity, and allosteric inhibitors inhibit the enzyme’s activity. Thus, the binding of modulators to the regulatory site alter the kinetic characteristics of an allosteric enzyme.
- Substrate binding to the catalytic site of an allosteric enzyme is cooperative, which means binding of a single substrate molecule shifts the equilibrium from the enzyme’s inactive state (T-state) to an active state (R-state) and promotes binding of another substrate.
- Modulators are inhibitory or stimulatory in function. Allosteric modulation refers to the conformational changes in the enzyme or alternations in the enzyme’s activity after the binding of effectors to the allosteric site. Effectors are non-substrate molecules that non-covalently bind to the enzyme’s regulatory site and positively or negatively affect the reaction rate.
Types
The allosteric inhibitors and activators attach to the T-form and R-form, respectively. Thus, based on the function of modulators, there are two types of regulation.
- The homotropic-modulator serves as a substrate and regulatory molecule that activates the enzyme’s activity in homotropic regulation.
- Oppositely, heterotropic-modulators only function as the regulators that may either behave as activators or inhibitors in heterotropic regulation. Therefore, substrates and modulators are different entities in heterotropic regulation.
Positive modulation is also called allosteric activation. Allosteric activators cause remodelling of the enzyme’s active site so as to increase the affinity for substrate binding. It drastically increases the reaction rate. Negative modulation is also called allosteric inhibition. Allosteric inhibitors alter the enzyme’s active site but decrease the affinity for substrate binding.
Models of Allosteric Regulation
Allosteric enzymes exist in both active (R-state) and inactive (T-state) states. Based on this aspect, there are two types of allosteric models.
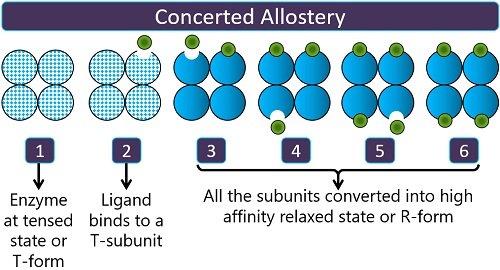
Symmetry allostery: It was introduced by Jacques Monad and his associates in 1965, and it is also known as the concerted model. Here, the binding of one ligand to the T or tense subunit converts all the other subunits to R or relaxed state. Thus, it causes the overall conversion of the tense subunit to a high affinity relaxed form.
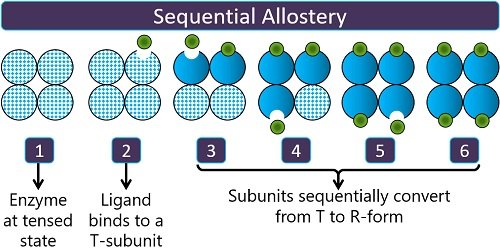
Sequential allostery: It was explained by Koshland Jr. in 1966. One substrate or ligand primarily binds to the enzyme’s specific site or “T-subunit” and results in a conformational change in the neighbouring subunits one by one. Thus, it causes the sequential conversion of subunits from T to R state.
Sigmoidal Curve by Allosteric Enzymes
Allosteric enzymes are the regulatory proteins that disobey the Michaelis- Menten kinetics due to multiple active sites and subunits. Instead, they follow a sigmoidal curve. The concentration of modulators and substrates greatly influence the kinetic behaviour of allosteric enzymes.
With allosteric enzymes, the binding affinity and catalytic activity of substrate or ligand to a single subunit can alter other subunits’ properties of the same enzyme. Such interaction results due to a cooperative property of allosteric enzymes, i.e. the binding of substrate to an enzyme’s catalytic site affect the binding of substrate to other active sites as well.
The cooperative binding of substrates to the respective subunits of an allosteric enzyme accounts for the sigmoidal curve of reaction velocity versus the substrate concentration, as you can see in the diagram below.
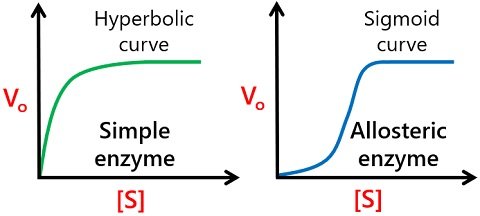
The sigmoidal curve has three significant stages, namely exponential, linear and plateau. First, the reaction rate increases exponentially due to an increase in substrate concentration. Then, the relationship between the enzyme and substrate switches to a linear phase, where more substrates are likely to bind with the respective sites.
The T and R state equilibrium depends on the concentration of the substrate. More enzymes are found in the R state at high [S], whereas the T state exists in insufficient substrate concentration. However, once the enzyme becomes fully saturated, the substrate concentration does not influence the reaction rate.
Conclusion
Therefore, allosteric regulation involves the association of the effector molecule with the enzyme’s allosteric site. Effectors can positively or negatively affect the enzyme’s activity. The regulation of allosteric enzymes follows the cooperative phenomenon and accounts for a sigmoidal curve.