Excision repair is one of the types of DNA repair systems. It is an enzymatic process, which follows a cut, copy and paste mechanism. A cutting mechanism involves removing a damaged part or unusual bases by specific enzymes like glycosylases and helicases. Then, a copying mechanism uses DNA polymerase.
DNAP-I uses the template DNA to form a new strand from the 3′-OH primer site, which eventually replaces the damaged portion of the DNA. At last, the pasting mechanism makes the use of DNA ligase to paste the disjointed DNA fragments. After a cut, copy and paste process, DNA appears to be fully intact and repaired.
Excision repair is named because it is a type of DNA repair mechanism in which the unwanted bases are directly excised by the combination of enzymes and replaced by the new DNA. The excision repair can be employed for different lengths like a very short, short and long patch of DNA.
It involves two methods: BER (base excision repair) and NER (nucleotide excision repair). A very short patch DNA refers to a single unwanted base in the DNA, and a base excision repair system repairs it. A short and long patch DNA refers to more than one unwanted bases in the DNA, and it is repaired via a nucleotide excision repair system.
Content: Excision Repair
Definition of Excision Repair
An excision repair refers to the DNA repair mechanism that deals with the damaged part of the DNA by excising either a single unwanted base or nucleotides with the new DNA bases. Excision repair uses different enzymes detach and repair the mutated or the damaged part of the DNA.
Types
Excision repair involves two methods, depending upon the excision mechanism:
Base Excision Repair
BER refers to the excision repair mechanism, which removes the small base adducts in DNA. In simple words, it is a type of short patch excision repair mechanism.
This repair system does not distort the configuration of the DNA helix. BER mainly repairs the mutated DNA caused by the endogenous mutagens. Base excision repair can correct the chemical changes in the DNA.
Hydrolysis and oxidation are the two leading causes of DNA damage. Hydrolysis cause depurination or depyrimidination in the DNA, which ultimately forms an AP-site. The AP-site will indicate the damaged part of the DNA.
BER can also cause deamination of cytosine into uracil, which leads to the transition mutation in the DNA. Oxidation changes the guanine base to 8-Oxo-G (pairs with A instead of C) and results in the formation of two daughter duplexes (8-Oxo-G: A and T: A).
Requirements
BER uses a combination of enzymes like DNA glycosylases, AP-endonuclease, DNA polymerase-I and DNA-ligase.
- DNA glycosylases: Its function is to recognize the unwanted base pairs and the AP-sites in the DNA.
- AP-endonuclease: Its function is to create a nick in the AP-site or damaged part of the DNA.
- DNA polymerase-I: It performs a functional role in the base excision repair by performing two activities. Through first, it cut out the AP-site via 3’-5’ exonuclease activity. Secondly, it synthesizes new DNA base pairs via 5’-3’ polymerization activity to fill the gap created by the AP-endonuclease.
- DNA ligase: It seals the gap and joins the DNA fragments. DNA ligase-I with the cofactor (XR C1) seals the gap in short patch BER.
Process
In BER, unwanted bases like uracil, hypoxanthine, 3-methyl adenosine occur in the base sequence of DNA that results in a chemical change. Uracil is not a part of DNA, but it can pair with the usual DNA sequence that leads to a transitional mutation.
The incorporation of hypoxanthine in the DNA will mislead the DNA for the synthesis of some essential proteins. Similarly, 3-methyl adenosine causes replication errors. The process of base excision repair involves the following steps:
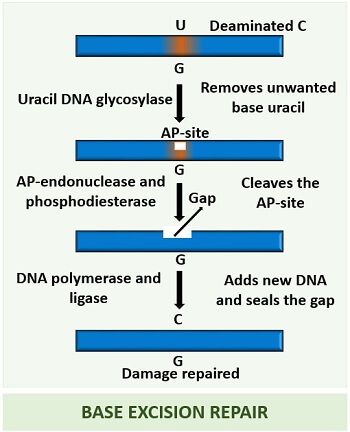
Firstly, glycosylases recognize the presence of unwanted bases in the DNA and eventually cleaves that section. The enzyme glycosylases only remove the unwanted bases, not the DNA backbone.
After the action of glycosylase, a gap appears at the site from where an unwanted base has been removed. This site will be called “AP-site” or an apurinic/apyrimidinic site. An AP-site represents the damaged part of the DNA. Thus, an AP-endonuclease enzyme will create a nick in the damaged section.
Then, a DNA polymerase-I cleaves the AP-site through its 3’-5’ exonuclease activity. Besides exonuclease activity, it also fulfils the gap created by the AP-endonuclease by adding new base pairs through its 5’-3’ polymerization activity. At last, DNA ligase seals the gap between the DNA fragments.
Nucleotide Excision Repair
NER refers to the excision repair mechanism, which removes the long base adducts in DNA or the whole nucleotide sequence. It is a type of long patch excision repair mechanism.
It distorts the configuration of the DNA helix. NER mainly repairs the DNA damage that originated due to the exogenous mutagens. NER repairs the damage induced by the UV-light in a DNA, oxidative damage and large chemical adducts.
Requirements
NER makes the use of Uvr-BC, Uvr-D and DNA polymerase-I and DNA-ligase enzymes.
- Uvr-BC: It is the multisubunit enzyme complex, where the Uvr-BC complex is encoded by the uvr-B and uvr-C genes, respectively. Uvr-BC recognizes the damaged DNA segment induced by the UV-radiation. It associates with the T=T dimer.
- Uvr-D: It is a helicase enzyme, which unwinds the DNA and chops the damaged DNA segment.
- DNA polymerase-I: It fills the gap by synthesizing new DNA base pairs through its 5’-3’ polymerization activity.
- DNA ligase: It seals the gap between the DNA fragments.
Process
In NER, the formation of pyrimidine dimer occurs in the base sequence of DNA due to physical changes or UV-rays. The method of nucleotide excision repair involves the following steps:
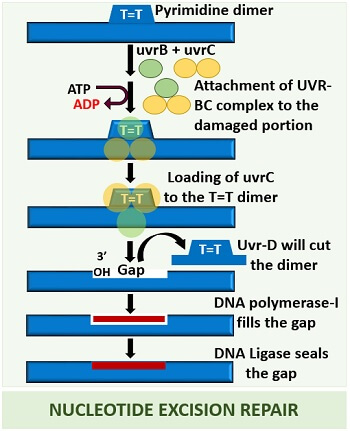
A thymine=thymine dimer forms as a result of potent UV-radiation. Uvr-B enzyme unites with the Uvr-C and forms a Uvr-BC complex to remove the dimer. The formation of the Uvr-BC complex gets energy by the hydrolysis of ATP molecule. Uvr-BC subunit scans and recognizes the damage in the DNA segment.
Then, the Uvr-B enzyme loads the Uvr-C enzyme and functions as an endonuclease enzyme. Thus, the Uvr-C enzyme forms a cleavage site for the further processing of DNA. This process also requires the utilization of energy.
After that, Uvr-D binds to the cleavage site and unwinds or cleaves the damaged DNA segment. An enzyme Uvr-D works like helicase-II, as it performs unwinding of the mutated portion. It utilizes ATP.
Finally, a DNA polymerase-I fulfils the gap created by the Uvr-D helicase enzyme by adding new DNA bases through its 5’-3’ polymerization activity. At last, DNA ligase seals the gap between the DNA fragments.
It is a very good note. I get the information that i want to know about DNA excision repair.