Bacterial transcription occurs in the cytoplasm. Transcription merely refers to the mechanism of mRNA synthesis complementary to the DNA template strand. The whole process is regulated enzymatically by the RNA polymerase holoenzyme. The study of transcription in prokaryotes is quite simple than the eukaryotic transcription. Besides RNA polymerase, prokaryotic transcription uses some protein factors that direct the mRNA synthesis.
Transcription is the first stage of gene expression, synthesizing linear mRNA from the DNA specific sequences. It uses a template DNA strand to synthesize RNA with a similar base sequence of the coding DNA strand. The difference between template and coding strand is that the former moves in a 3′-5′ end and the latter moves in a 5′-3′ direction.
Bacterial transcription only produces short stretches of RNA that serve as the primary transcript. Initiation, elongation and termination are the three typical stages involved in transcription. This post describes the definition, diagrams and steps of bacterial transcription.
Content: Bacterial Transcription
Definition of Bacterial Transcription
Bacterial transcription is the first stage of gene expression that occurs in the bacterial cell cytoplasm. It only needs RNA polymerase holoenzyme to synthesize all the types of RNA (mRNA, tRNA and rRNA). Additionally, protein factors like sigma and rho protein are necessary to regulate transcription.
Bacterial transcription typically involves three stages abortive initiation, elongation of mRNA and stalling of RNA polymerase by intrinsic RNA sequences or rho factor. Primarily, specific binding between RNA polymerase and the promoter sequences is necessary. Before transcribing mRNA, the unwinding of the DNA helix by the RNA polymerase becomes necessary near the gene that is getting transcribed.
As a result, a transcription bubble forms with two exposed DNA strands (coding and template strand). Transcription only utilizes the template DNA strand to produce RNA in the 5’ to 3’ direction by adding nucleotides one by one to the free 3’-OH end. The RNA polymerase dissociates via intrinsic or rho-dependent termination after transcribing 10-12 bps long RNA.
Key Terms
Central dogma: According to the current bioinformatics concept, transcription is the first stage of gene expression where the genome is transcribed into the transcriptome by the DNA-directed RNA polymerase. The genome is the total DNA contained in an organism. The transcriptome is the RNA or the initial product required to synthesize proteins or serves as active protein-coding genes.

Prokaryotic RNA polymerase is the core enzyme possessing catalytic property. But, it functions as the holoenzyme along with the sigma factor, i.e. RNAP cannot be considered the holoenzyme without the sigma factor. Unlike eukaryotic transcription, bacterial transcription only uses RNA polymerase holoenzyme to synthesize mRNA, tRNA and rRNA. It possesses a core complex with five subunits (ββ′α2ω).
- Two α-subunits assemble the RNAP on the DNA.
- β-subunit adheres with the ribonucleoside triphosphate of nascent mRNA.
- β′-subunit attaches to the DNA template strand.
- The function of the ω-subunit is not known.
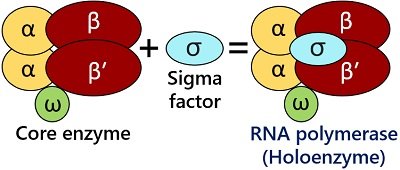
Sigma protein is the sixth subunit that assembles during the gene transcription and dissembles during the transcription termination. It associates with the core complex of the RNA polymerase to make it biochemically active. It aids in promoter recognition, correct binding of the RNA polymerase to the promoter sequences of the DNA and promotes DNA-unwinding at the start site. A sigma protein subunit dissociates after transcription initiation.
Rho protein possesses helicase activity that dissociates RNA polymerase and regulates transcription termination in prokaryotes.
Steps of Bacterial Transcription
It involves the following stages depicted in the diagram below.
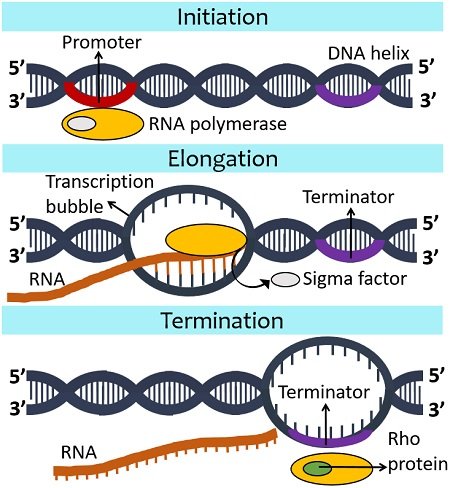
Initiation
It is the primary stage of transcription. Here, the RNA polymerase binds to the specific sequence or promoter region of the coding DNA strand. The majority of bacteria have two consensus sequences (-10 and -35) in the promoter region.
- The -35 sequence is the promoter’s recognition site, possessing six nucleotide bases (TTGACA) and present about 35 bases away from the initiation site.
- The -10 sequence (Pribnow box or TATA box) comprises six nucleotide bases (TATAAT), which is present ten bases upstream from the start or +1 site.
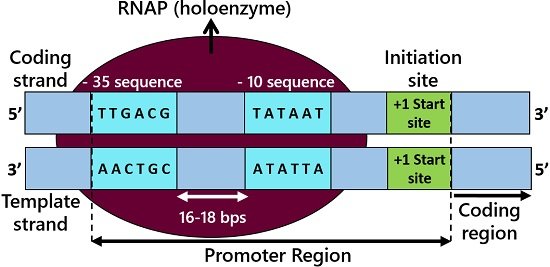
Therefore, promoters generally exist upstream of the transcription initiation. Firstly, the RNA polymerase assembles at the promoter region. But, the association of RNA polymerase with the promoter region is transient.
RNA polymerase is the core enzyme that alone cannot initiate transcription or recognize the promoter region. RNAP needs a sigma factor to function as a holoenzyme or complete enzyme. A sigma factor or subunit binds to the core of RNAP. It plays a crucial role in recognizing the consensus sequences in the DNA and promoting specific binding between RNAP and the promoter region.
Afterwards, the RNAP slightly modifies its shape or generally stretches itself (upto 60-80 base pairs) to reach the promoter region because, at the very start, RNAP adheres far away from the promoter site.
Abortive Initiation
- Primarily, a closed binary complex forms by the association of RNAP holoenzyme and the promoter sequence. Transcription in prokaryotes involves several abortive cycles to synthesize short mRNA transcripts.
- A closed complex results in partially melting or unwinding of the DNA. Therefore, an open binary complex forms by the tight binding between the RNAP holoenzyme and unzipped DNA.
- Then incorporation of two ribonucleotides and the formation of a phosphodiester bond occurs between them. As a result, an initiation ternary complex forms.
- In the promoter clearance stage, the core RNA polymerase leaves the promoter site and transits to the elongation phase of transcription. Finally, the sigma factor dissociates.
Elongation
It is the second stage of transcription. Here, the RNA polymerase reads the template DNA strand and forms the primary transcript, i.e. mRNA. Elongation of the primary transcript occurs in the direction of 5’ to 3’ end. Thus, the RNA is complementary or antiparallel to the DNA template or anti-sense strand.
The base sequence of newly formed RNA is similar to DNA coding or positive sense strand (except for the nitrogenous base uracil). During elongation, RNA polymerase synthesizes RNA through one of its catalytic properties. Unlike DNA polymerase, RNAP does not need primer. Elongation of the ternary complex involves the following stages:
- DNA unwinds as the elongation proceeds due to breaks in hydrogen bonds connecting the complementary base pairs. Base pairing between the DNA and transcribing RNA is not stable. However, the RNAP serves as a stable linker.
- Nucleotides combine to the 3’-OH end of the RNA. The free 3’-end of the growing RNA facilitates a nucleophilic attack on the α-phosphate of the incoming nucleoside triphosphate.
- Protein bridge allows the addition of a single nitrogenous base to the growing chain by adding more specificity. It generally synthesizes short strands of RNA (10-12 base pairs long).
Termination
Rho-independent and rho-dependent are the two mechanisms regulating transcription termination. Rho-independent or intrinsic termination uses terminator sequences within the RNA. Here, the palindromic sequences in the newly synthesized serve as the terminator sequences that signal the RNA polymerase to terminate the transcription.
The palindromic sequences in the RNA produce stem and hair loop structure due to GC-rich regions. RNAP dissociates from the DNA template strand, thereby terminating transcription. Conversely, a rho protein directs the rho-dependent termination. It is the hexameric helicase enzyme that binds to the rho-utilization site or rut-site of the growing RNA.
Rho runs along the path of growing RNA towards the RNA polymerase, thereby dissociating RNAP and destabilizing the interaction between the template and mRNA. Finally, the transcription product, i.e. mRNA released from the elongation complex, is generally inactive. It goes through some post-transcriptional modifications to become functionally active.
Conclusion
We can conclude that bacterial transcription is relatively simpler as it uses a single RNA polymerase and does not require additional transcription factors. Only the template strand is used to make RNA copies, and we could say that the transcription mechanism is selective. The transcription product or working RNA copies express the genetic information of the template DNA.